Unnesting unsplittable strings
Here’s another quick post about turning human-readable data into something we can actually work with.
For my research, I was cleaning some data about rodent specimens from different sites examined in this publication by Araujo Fernandes et al (2009). Once I had managed to read the table into R, I recognized the various little tricks commonly used to condense information and save space on a table without losing meaning.
This is what the first few entries of the table look like.
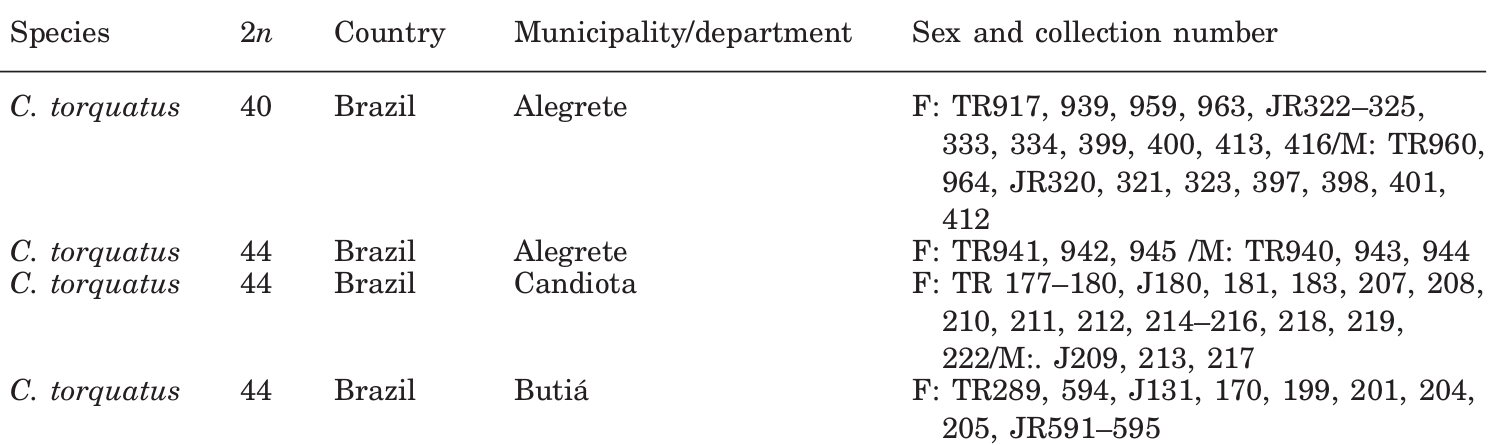
We are interested in the last column “Sex and collection number”. Two different variables have been condensed in a single column, but that’s fine because there are consistent delimiters and separators. Also, consecutive numbers have been shortened to imply a sequence.
The trickiest part are the specimen ID values. These are made up of an acronym and a number, and in this case the collection IDs appear as wrapped inline text for each sampling location. When specimens from a single location come from more than one collection, there is no explicit separator, but we can tell that the ID numbers correspond to the collection that precede them until we see a new acronym that implies otherwise. None of this is explained in the table caption, but this is a common convention and it’s mostly self-explanatory.
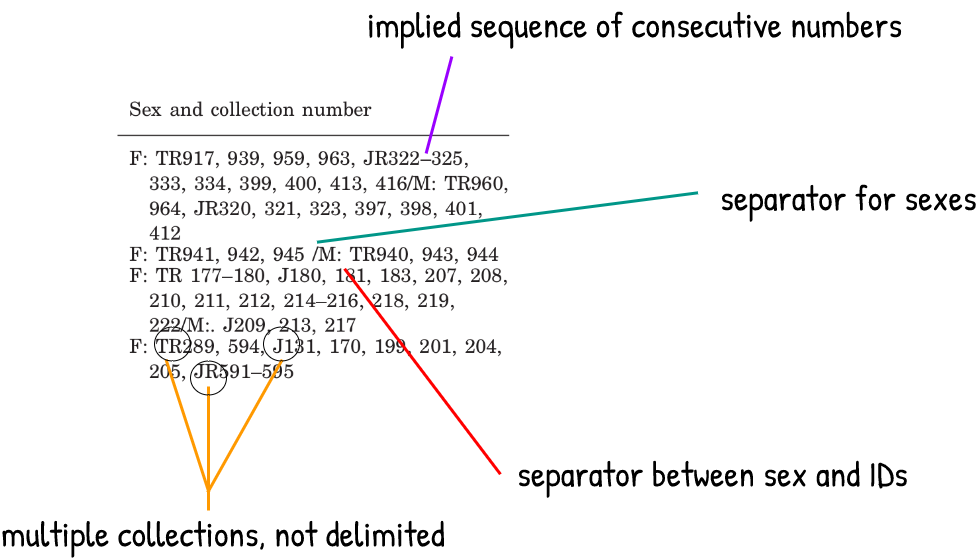
The non-delimited collection acronyms are the obvious challenge here. I had encountered this issue before, but I always gave up and solved this through laborious manual editing. Now that I found an adequate solution, I’m using this post to share it and to document my progress with using tidyr and with writing regular expressions (instead of my previous method of blindly copying regex from StackOverflow and hoping it works).
Let’s load the required packages and set up a subset of the data for this demo:
library(dplyr)
library(tidyr)
library(stringr)
library(purrr)
dat <- tibble::tribble(
~municipality, ~sex_and_collection_number,
"Alegrete", "F: TR917, 939, 959, 963, JR322–325, 333, 334, 399, 400, 413, 416/M: TR960, 964, JR320, 321, 323, 397, 398, 401, 412",
"Alegrete South", "F: TR941, 942, 945/M: TR940, 943, 944",
"Candiota", "F: TR 177–180, J180, 181, 183, 207, 208, 210, 211, 212, 214–216, 218, 219, 222/M: J209, 213, 217",
"Butiá", "F: TR289, 594, J131, 170, 199, 201, 204, 205, JR591–595",
"Candelária", "M: MNHNA1885",
"Cachoeira do Sul", "F: TR921–924"
)First we can put the values of the sex variable into their own column, by splitting and unnesting and then separating on the existing delimiter.
# split and unnest sexes
dat <- dat %>% unnest(sex_and_collection_number = strsplit(sex_and_collection_number, "\\/"))
# separate
dat <- dat %>% separate(sex_and_collection_number, into = c("sex", "specimen"), sep = ": ")We’re making progress, but we cannot do an further unnesting until we deal with the rows that have more than one collection.
> dat
# A tibble: 9 x 3
municipality sex specimen
<chr> <chr> <chr>
1 Alegrete F TR917, 939, 959, 963, JR322–325, 333, 334, 399, 400, 413, 416
2 Alegrete M TR960, 964, JR320, 321, 323, 397, 398, 401, 412
3 Alegrete South F TR941, 942, 945
4 Alegrete South M TR940, 943, 944
5 Candiota F TR 177–180, J180, 181, 183, 207, 208, 210, 211, 212, 214–216, 218,…
6 Candiota M J209, 213, 217
7 Butiá F TR289, 594, J131, 170, 199, 201, 204, 205, JR591–595
8 Candelária M MNHNA1885
9 Cachoeira do Sul F TR921–924 This is where the regex magic comes in, we can use a lookbehind and a backreference in the replacement argument to insert a new delimiter before all the instances of uppercase characters at a word boundary (i.e. the collection acronyms).
# note the negative lookbehind, and the backreference in the replacement
dat <- dat %>% mutate(specimen = str_replace_all(specimen, "((?<!^)\\b[A-Z])", ";\\1"))If we inspect the first few rows, we can see the inserted semicolons that now delimit the different collections.
municipality sex specimen
<chr> <chr> <chr>
1 Alegrete F TR917, 939, 959, 963, ;JR322–325, 333, 334, 399, 400, 413, 416
2 Alegrete M TR960, 964, ;JR320, 321, 323, 397, 398, 401, 412 We can now unnest the collections using these new delimiters
# unnest collections
dat <- dat %>% unnest(specimen = strsplit(specimen, ";")) %>% mutate(specimen=str_squish(specimen))I recommend this book by Michael Fitzgerald as a handy guide to regex. Important concepts are well explained, and it has a bat on the cover.

Afterwards, we can put the collection acronyms into a new variable and unnest the specimens (this part is a little hacky but it works).
#extract acronyms
dat <- dat %>%
mutate(museumAbbr = str_extract(specimen, "^[A-Z]+")) %>%
mutate(specimen = str_squish(str_remove(specimen, "^[A-Z]+")))
# trailing commas
dat <- dat %>% mutate(specimen=str_remove(specimen,",$"))
# unnest specimens
dat <- dat %>% unnest(specimen = strsplit(specimen, ",")) %>%
mutate(specimen=str_squish(specimen))The first few rows now look like this:
> dat
# A tibble: 53 x 4
municipality sex museumAbbr specimen
<chr> <chr> <chr> <chr>
1 Alegrete F TR 917
2 Alegrete F TR 939
3 Alegrete F TR 959
4 Alegrete F TR 963
5 Alegrete F JR 322–325
6 Alegrete F JR 333 The last issue are the series specimens with consecutive ID numbers. We need to expand these series before unnesting. I wrote a little function to do this and vectorized it to work inside mutate. Once the series are expanded into a delimited string we can finally unnest the specimens.
# for consecutive specimens
expand_series <- function(ser_vector, separator) {
words <- str_split(ser_vector, paste(separator))
if (length(words[[1]]) == 1) {
return(ser_vector)
} else {
start_str <- pluck(words, 1, 1)
}
end_str <- pluck(words, 1, 2)
paste0(seq(as.integer(start_str), as.integer(end_str)), collapse = ", ")
}
expand_series <- Vectorize(expand_series)
# expand
dat <- dat %>% mutate(specimen=expand_series(.$specimen,'–'))
# unnest specimens again
dat <- dat %>% unnest(specimen = strsplit(specimen, ", ")) %>%
mutate(specimen=str_squish(specimen))Now we can work with the 68 specimens originally mashed into six rows, one for each of the municipalities. We can get some summary data easily with this long-form and tidy data model. For example: a tally of how many specimens of each sex there are from each location.
> dat %>% count(municipality,sex)
# A tibble: 9 x 3
municipality sex n
<chr> <chr> <int>
1 Alegrete F 14
2 Alegrete M 9
3 Alegrete South F 3
4 Alegrete South M 3
5 Butiá F 13
6 Cachoeira do Sul F 4
7 Candelária M 1
8 Candiota F 18
9 Candiota M 3That’s all. Let me know if you have any feedback or questions.

