This post was discussed in the Rweekly Highlights Podcast (Episode 118). Listen here.
From small one-off scripts to massive interactive apps, most workflows use packages that help us extend the capabilities of a programming language. Sometimes we only need some example data or a few additional functions. Other times we want to add grammars, data structures, printing methods, graphical devices, or OOP programming systems.
Rather than only working interactively, it helps to use scripts and save our source code for repeatability and transparency. When we need something from a package that fits our needs, we generally call this package from a script, so properly documenting how we use packages within scripts can make code easier to understand, debug, and share.
Can you figure out which packages here don’t actually exist?
# packages I may or may not need for this task
library(e1071)
library(yulab.utils)
library(wk)
library(windex)
library(xonkicks)
library(whisker)
library(labbo)
library(stringx)
library(castor)One easy way to add information about the packages we use is with code comments for package load calls. In R, anything after the commenting symbol “#” will be ignored during evaluation. It’s best practice to not overuse comments and to write self-explanatory code, but recording certain details about the packages we’re calling in a script can be useful.
The code above can be easily enhanced with the title and source for each package (as installed on my machine), and this is potentially helpful.
# packages with title and source as comments
library(e1071) # Misc Functions of the Department of Statistics, Probability Theory Group CRAN v1.7-9
library(yulab.utils) # Supporting Functions for Packages Maintained by 'YuLab-SMU', CRAN v0.0.4
library(wk) # Lightweight Well-Known Geometry Parsing, CRAN v0.6.0
library(windex) # Analysing Convergent Evolution using the Wheatsheaf Index, CRAN v2.0.3
library(xonkicks) # not installed
library(whisker) # for R, Logicless Templating, CRAN v0.4
library(labbo) # not installed
library(stringx) # Drop-in Replacements for Base String Functions Powered by'stringi', [github::gagolews/stringx] v0.2.4
library(castor) # Efficient Phylogenetics on Large Trees, CRAN v1.7.8The two made up packages were xonkicks and labbo.
To easily create code comments about the package load calls in a script, we can use the annotater package (read more about the package here). The functions build code comments using information already supplied by a package in its DESCRIPTION file or within its internal lists of functions and bundled datasets. This can be quite helpful for sharing code online, teaching, or making sense of existing scripts.
At present, annotater can add the following details to package load calls in scripts or markdown (Rmd/Qmd) files:
- package title
- package version
- package repository source (e.g. CRAN, GitHub, Bioconductor, etc.)
- which functions from each package are being used
- which datasets from each packages are being used
The number of possible package annotations has grown thanks to feedback and community contributions, but I still had a pressing question:
What are people commenting about their loaded packages?
With so much public code available, it is now possible to search for .R, .Rmd and .Qmd files online, look for package load calls (e.g., library(data.table)), and then check for code comments after these lines (e.g., library(data.table) # using dev version).
A good source of data for this can be the weekly GitHub snapshots available on Google’s BigQuery cloud platform. This 3TB+ dataset includes the content of 163 million files, all searchable with regular expressions.
This post by Vlad Kozhevnikov explains how to write SQL for BigQuery to select the IDs of all relevant files and then select content of R files. After that we can run another query to search the script contents for code comments after library() calls.
The queries for .R files look like this (change with your respective BigQuery project, dataset, and table names). I repeated this for .R, .Rmd, and .Qmd files.
-- Find all files with .R extension (case insensitive)
SELECT *
FROM `bigquery-public-data.github_repos.files`
WHERE lower(RIGHT(path, 2)) = '.r'
-- Match to get file contents
SELECT *
FROM `bigquery-public-data.github_repos.contents`
WHERE id IN (select id from `YOURBQprojectID.yourDATASET.yourTABLE`)
– Regex match library calls with a comment after whitespace (except for newlines)
SELECT *
FROM `YOURBQprojectID.yourDATASET.yourTABLE`
WHERE REGEXP_CONTAINS(content, r'library\(.*\)[ \t]?#')Many thanks to fellow instructors Steve Condylios and Amit Kohli for pointing me in the right direction!
My latest query found 500,641 .R files, 44,839 Rmd, and 242 Qmd files in the GitHub Data snapshot (March 14, 2023) of which 3968 included at least one code comment after a library() call.
A good proportion of these 545,722 files won’t be scripts that people are meant to see or interact with (for example, .R files in Shiny apps). Still, the 0.7% of files with comments about the loaded packages will have some interesting information about what us as users are commenting.
Also following Vlad’s post, I was able to connect with BigQuery from R using bigrquery and download the tables to objects in my R environment. Once in R, there is some cleaning and parsing to get each file’s library calls and respective comments (if any) into a tidier structure.
All the R code for this post is at the Gist at the end in case anyone is interested. This approach uses regex to parse code but also other cool functions to treat code as code rather than strings. In short, I had to:
- get only the lines with library calls in the field that stored all the script’s content
- split each call into a separate row
- separate comments into their own variable
- clean up spaces and unmatched brackets (from wrapped
library()calls) - parse the calls and extract the first argument of the
library()calls to get package names and not other arguments people sometimes use. - various summary statistics
General overview
For the 3968 (out of >500,000 files) that included at least one code comment after a library load call, the mean number of packages loaded was 6 per script (minimum 1, maximum 203!) and across these scripts over half (55%) of the packages mentioned in each one have a code comment.
Let’s see a set of commented calls sampled at random:
| pkgname | comment |
|---|---|
| roxygen2 | If not availble install these packages with ‘install.packages(…)’ |
| tidyverse | load c(dplyr, tidyr, stringr, readr) due to system doesn’t work. |
| scales | install.packages(“scales”) |
| GenomicRanges | Required for defining regions |
| devtools | session_info |
| magrittr | Pipes |
| forecast | varios metodos para pronóstico |
| gridExtra | for multiple plots on a page |
| car | using external libraries |
| rgl | Need to install this package first |
| reshape2 | For converting wide to long |
| MplusAutomation | for extracting gh5 |
| ggplot2 | for plotting color plots with legend |
| ggplot2 | For plotting |
| DataCombine | for the slide function |
| irr | fleiss’ kappa |
| corpcor | for fast computation of pseudoinverse |
| ggplot2 | For plotting |
| dplyr | alternative, this also loads %>% |
| scales | For formating values in graphs |
Now let’s see the comments that repeat the most regardless of which package they’re meant for:
| comment | n |
|---|---|
| Pipes | 206 |
| enables piping : %>% | 93 |
| For graphing | 84 |
| data manipulation | 49 |
| for data manipulation | 38 |
Even with this small sample we can already see some patterns and shared themes in these comments. For example:
- Notes about installation
- What the package is being called for
- Which function or functions from the package are being used
- Pipes
Skimming the comments, a decent proportion of them are describing what a package was used for in general or mentioning the functions or datasets of interest. 22% of all comments start with the words “for”, “para”, and “pour” (case insensitive). Here’s another random sample:
| pkgname | comment |
|---|---|
| bcmaps | for BC regional district map |
| tmap | for maps |
| car | for initial parameter estimate |
| factoextra | for fviz_cluster(), get_eigenvalue(), eclust() |
| clusterSim | for Davies-Bouldin’s cluster separation measure |
| e1071 | Para la función svm |
| plotrix | for plotCI(), cld() |
| edgeR | for DGE |
| parallel | for examining machine |
| Boruta | for feature importance |
| missForest | for missForest() |
| grid | For ‘unit’ function in tick length setting |
| Amelia | for missmap() |
| tidyr | for data handling |
| MASS | For the data set # Use smoke as the faceting variable |
| Epi | for AUC evaluator |
| minpack.lm | for non-linear regression package |
| plyr | For the function “each” |
| MASS | for LDA |
| grid | for gList for likert plots w/ histograms |
~15% of comments had some kind of note about installation instructions or package source (matched the pattern “instal” or ‘CRAN’ or ‘github’).
We can even group the data by package find the most commented packages. For reference, these are the ten packages with the most comments (excludes duplicate comments):
| pkgname | n |
|---|---|
| ggplot2 | 208 |
| dplyr | 190 |
| plyr | 92 |
| MASS | 87 |
| tidyverse | 74 |
| reshape2 | 71 |
| stringr | 64 |
| scales | 64 |
| data.table | 60 |
| lubridate | 55 |
Many of these are tidyverse packages and sometimes the comments made note of that. I did not look at when these scripts are from, but they seem to cover and make note of important changes such as the melt/cast-gather/spread-pivot transition. See some comments for tidyr that mention reshaping in general:
| pkgname | comment |
|---|---|
| tidyr | for reshaping data (e.g., ‘gather’) |
| tidyr | instead of reshape2::melt |
| tidyr | key package for reshaping |
| tidyr | reshaping data |
| tidyr | library(Hmisc); library(reshape2); |
| tidyr | data reshaping |
| tidyr | because we’ll need to reshape the data |
| tidyr | For reshaping dataframes, specifically nesting |
| tidyr | this library contains the function that we’ll need in order to reshape the dataframe |
I had noted there are comments in different languages, so we can try to identify them with Google’s Compact Language Detector as implemented in the cld3 pacakge. The function is vectorized and easy to use.
| commentLanguage | n | percent | |
|---|---|---|---|
| en | 4820 | 0.72 | |
| no | 324 | 0.05 | |
| pt | 103 | 0.02 | |
| fr | 90 | 0.01 | |
| es | 88 | 0.01 | |
| ja | 85 | 0.01 |
The vast majority of annotations appear to be in English, followed by Norwegian, Portugese, French, Spanish, and Japanese. Because the comments are so brief and many include function names that aren’t real words, I highly doubt the accuracy some of these detected languages. I looked at some the comments that were supposedly in Norweigian and they were mostly short comments with function names and not words or sentences in any particular language. Still, I was interested in the Spanish-language comments and I found some similar to what I’ve written in the past. It’s pretty interesting to see how people describe packages.
| pkgname | comment |
|---|---|
| dplyr | paquete que veremos mañana y que por ahota lo necesitan otras funciones |
| dplyr | ya incluido en tidyverse |
| corrplot | grafico de correlacion |
| data.table | manipulacion de datos mayor rapidez que dplyr |
| NombreDelPaquete | Para usar el paquete |
| dplyr | Para cargar el paquete dplyr |
| stringr | Para la conversión de tipos numéricos a cadenas |
| psych | incluye describe() |
| IRdisplay | despliegue de resultados en jupyter para R |
| fdrtool | para half normal |
| Hmisc | algunas funciones de interes para describir datos |
| randomForest | construir enms |
| readxl | read_excel |
| stringr | creo que ni la usé |
| foreign | Solo para cargar el conjunto de datos de prueba en formato arff |
| ggplot2 | manipulacion de graficos - tidyverse |
| curl | Descargar desde internet |
I particularly like the comment “creo que ni la usé” or “I think I didn’t even use this package” because it makes the tools from annotater seem helpful. If I have a big script and am unsure whether or not I used a function or dataset from a package, the addins can check that for me.
As a quick exercise, here are seven comments sampled randomly for five of the packages in the data drawn as network graphs. I think that even without labeling the package name we could figure out what each one is based on the comments.
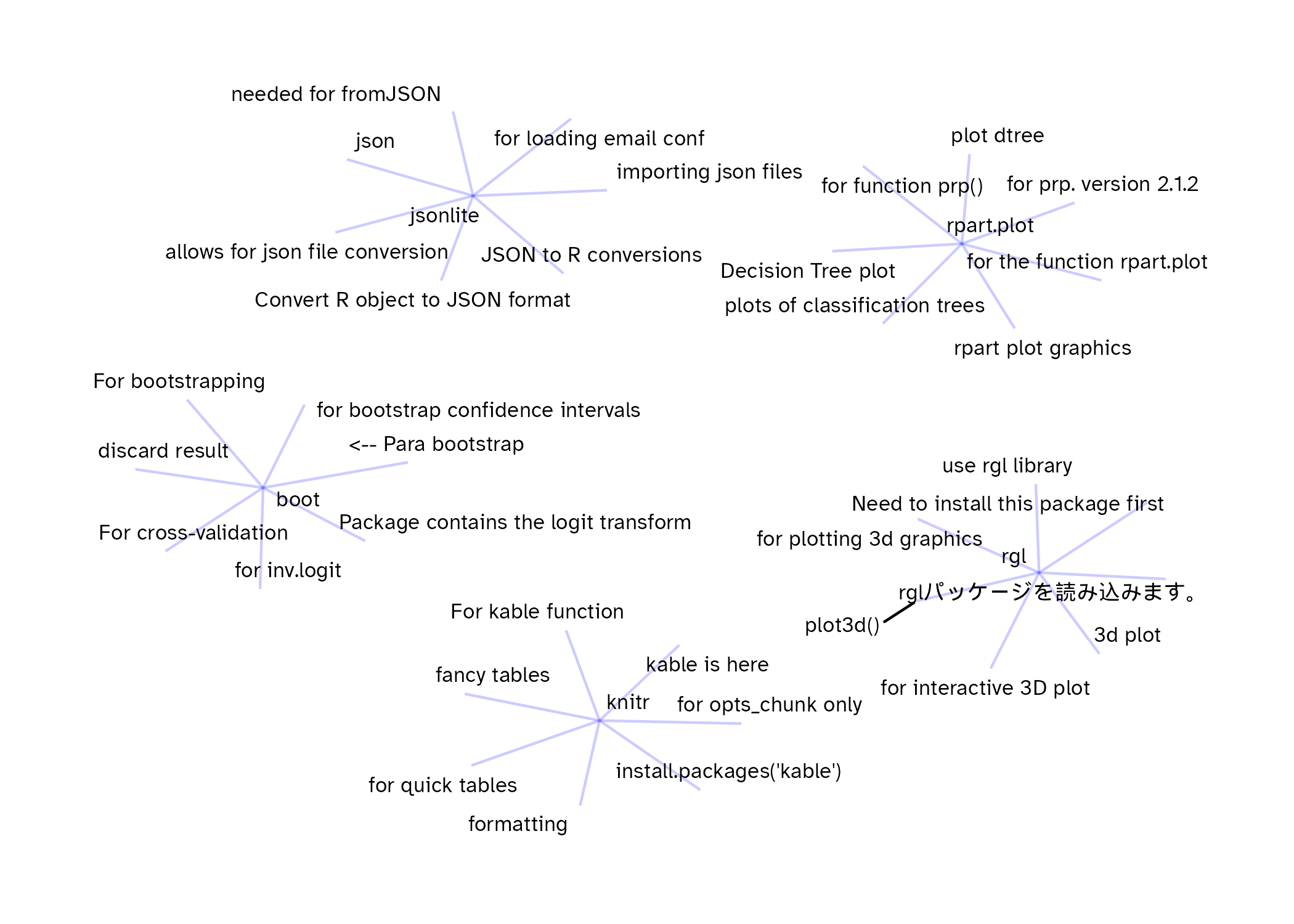
There is a lot to explore in these data (TidyTuesday potential??), but I’m quite happy with the overlap between what I found and the comments that can be created using the the tools that I’ve been working on.
Exploration code:
