Fixing broken and irregular column headers
Last week I saw Riva Quiroga masterfully import and clean a scary spreadsheet during an RLadies meetup, and I recognized some common issues with other people’s data for which I have already written relevant functions.
I also realized that I never wrote a dedicated post for the mash_colnames function in the unheadr package. This functionality has been around for a while now, but is worth describing in detail (plus I get to create some new teaching materials). Credit goes to Jarret Byrnes, who contributed the initial version of the function in this GitHub issue. I just added some tidyeval and a few enhancements for a better fit within the package. Internally, mash_colnames pivots the column headers and the first n rows of the data and ‘mashes’ whatever needs to be together columnwise.
mash_colnames() has two main uses. We can apply this nifty function to data with the following issues:
Data with the variable names split across > 1 rows (i.e. there are fragments of the headers in the first few data rows)
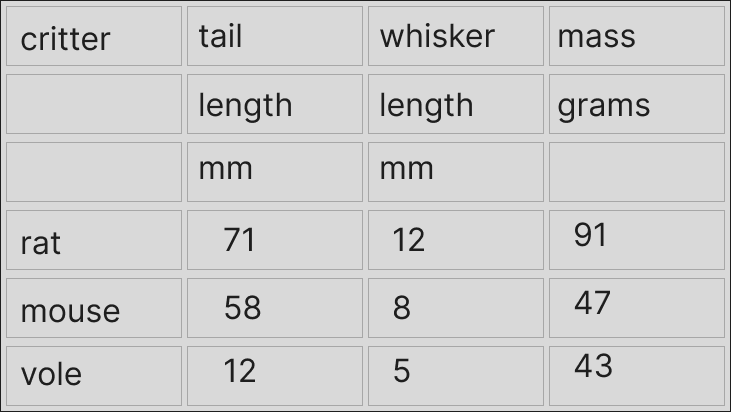
This little example here with rodent data has different bits of the column headers spread out ‘vertically’ for some reason. This is quite common, as far as I’ve seen. This next image below explains the problem a bit better. Notice that if there were any separators between the pieces of column headers, these are now implicit.
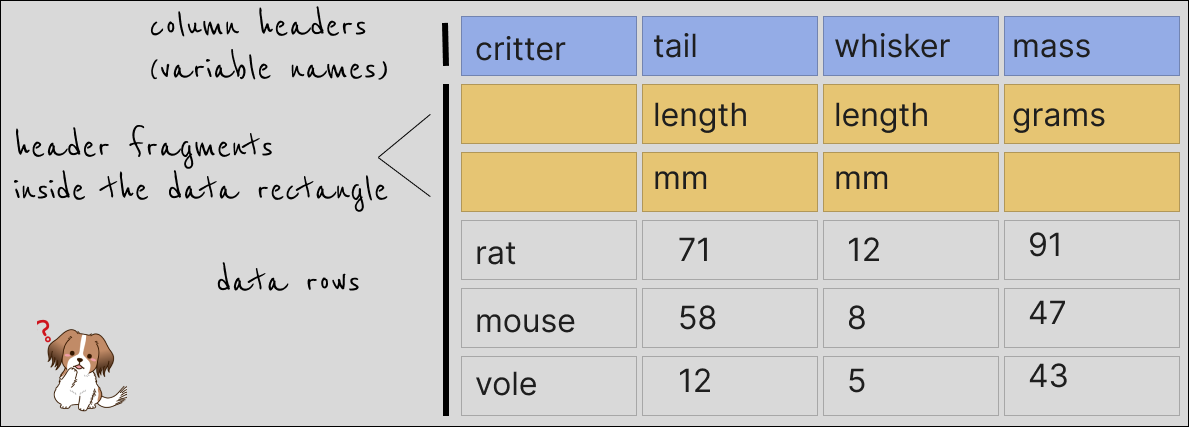
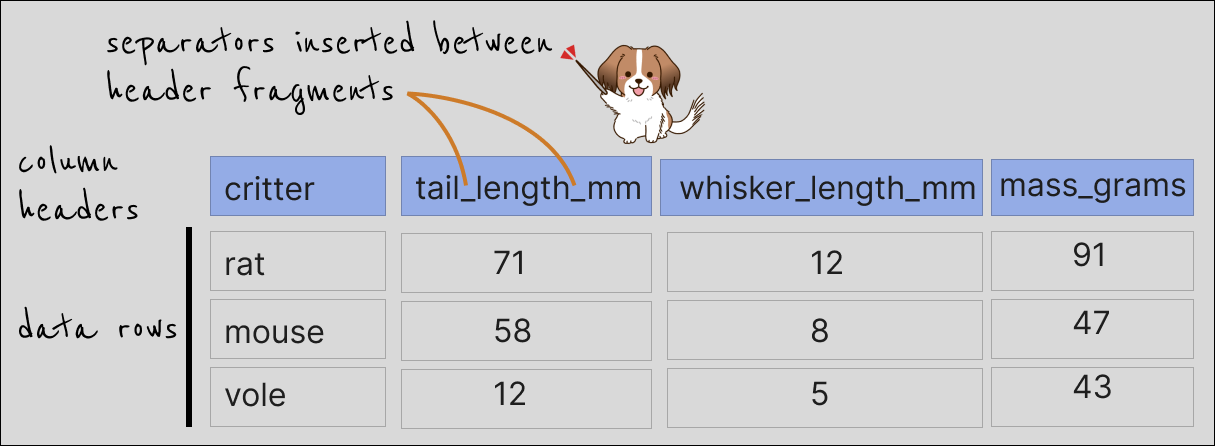
Ultimately, we most likely are interested in something like this:
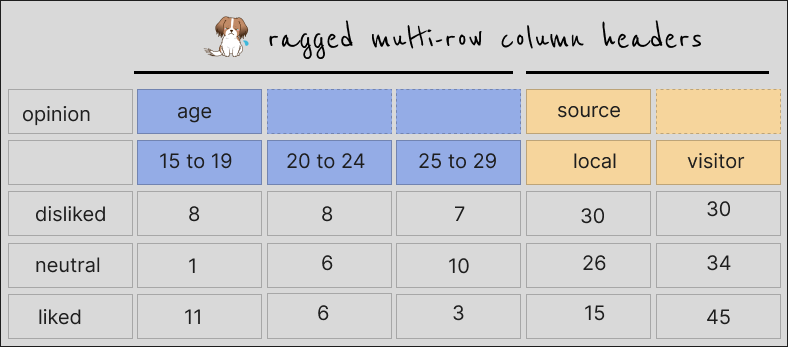
Names split across >1 rows but with gaps in the headers at the very top
This happens when cells were originally merged in a spreadsheet or formatted table, or maybe the gaps are there intentionally to imply that the values along this row are the same until a new one appears. I’m not sure about the correct terms for this but Charlie Hadley referred to this as “non-regular spanning of column headers”.
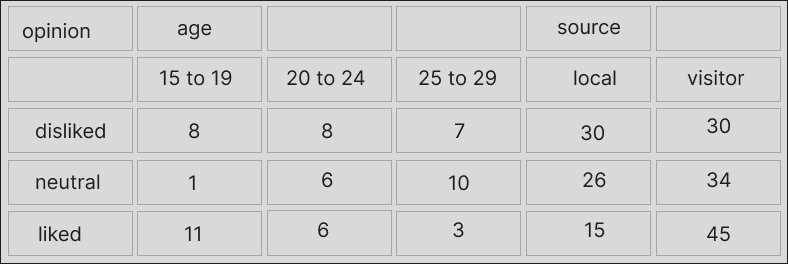
With colors to show which columns are meant to share a piece of header, the data look like this:
A more usable version of the data would look like this:
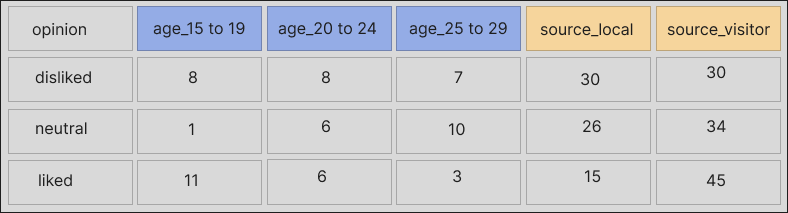
Having shown the two common issues that we can address with unheadr, let’s work through the same examples using code.
Working with code
Let’s set up the same example data from the images above and fix the issues.
library(unheadr) # CRAN v0.3.3
rodents <-
tibble::tribble(
~critter, ~tail, ~whisker, ~mass,
NA, "length", "length", "grams",
NA, "mm", "mm", NA,
"rat", "71", "12", "91",
"mouse", "58", "8", "47",
"vole", "12", "5", "43"
)The data in tibble form:
> rodents
# A tibble: 5 × 4
critter tail whisker mass
<chr> <chr> <chr> <chr>
1 NA length length grams
2 NA mm mm NA
3 rat 71 12 91
4 mouse 58 8 47
5 vole 12 5 43 Note the NAs padding the empty spaces.
To fix messy names broken across rows, we tell mash_colnames() how many data rows have header fragments in them. In this case, it’s two, the names don’t count as data rows. The default separator in the function is the underscore, but we can change it to spaces or dots or whatever.
rodents |>
mash_colnames(n_name_rows = 2) # underscore is the default separator The data with fixed names:
# A tibble: 3 × 4
critter tail_length_mm whisker_length_mm mass_grams
<chr> <chr> <chr> <chr>
1 rat 71 12 91
2 mouse 58 8 47
3 vole 12 5 43 Pretty nice! No more unnecessary NAs.
In some cases we may recognize messy names or similar issues in a dataset, so we skip the first row. It may look like this, with automated names that do not mean anything.
rodents_skip <-
tibble::tribble(
~X1, ~X2, ~X3, ~X4,
"critter", "tail", "whisker", "mass",
NA, "length", "length", "grams",
NA, "mm", "mm", NA,
"rat", "71", "12", "91",
"mouse", "58", "8", "47",
"vole", "12", "5", "43"
)> rodents_skip
# A tibble: 6 × 4
X1 X2 X3 X4
<chr> <chr> <chr> <chr>
1 critter tail whisker mass
2 NA length length grams
3 NA mm mm NA
4 rat 71 12 91
5 mouse 58 8 47
6 vole 12 5 43 For cases like these, we can use the keep_names argument to ignore the names when we’re mashing. In this case we work with three data rows, which hold all the pieces of the names.
Like so:
rodents_skip |>
mash_colnames(n_name_rows = 3, keep_names = FALSE)The result is the same as before.
# A tibble: 3 × 4
critter tail_length_mm whisker_length_mm mass_grams
<chr> <chr> <chr> <chr>
1 rat 71 12 91
2 mouse 58 8 47
3 vole 12 5 43 Ragged names
Let’s set up the example data with the gaps in the names column. When there are gaps in the column names we tend to skip the names during the import step. The keep_names argument really comes in handy here.
surveys <-
tibble::tribble(
~X1, ~X2, ~X3, ~X4, ~X5, ~X6,
"opinion", "age", NA, NA, "source", NA,
NA, "15 to 19", "20 to 24", "25 to 29", "local", "visitor",
"disliked", "8", "8", "7", "30", "30",
"neutral", "1", "6", "10", "26", "34",
"liked", "11", "6", "3", "15", "45"
)> surveys
# A tibble: 5 × 6
X1 X2 X3 X4 X5 X6
<chr> <chr> <chr> <chr> <chr> <chr>
1 opinion age NA NA source NA
2 NA 15 to 19 20 to 24 25 to 29 local visitor
3 disliked 8 8 7 30 30
4 neutral 1 6 10 26 34
5 liked 11 6 3 15 45 The approach is similar, and to deal with the gaps that imply a repeated value across columns, we use the sliding_headers argument. By setting it to TRUE we fill the gaps from left to right.
surveys |>
mash_colnames(2, keep_names = FALSE, sliding_headers = TRUE)The cleaned-up version is ready for further analysis.
# A tibble: 3 × 6
opinion `age_15 to 19` `age_20 to 24` `age_25 to 29` source_local source_visitor
<chr> <chr> <chr> <chr> <chr> <chr>
1 disliked 8 8 7 30 30
2 neutral 1 6 10 26 34
3 liked 11 6 3 15 45 That’s it! Feel free to reach out with any questions/feedback or if I was using the wrong data-structuring terms for this.

