Does anyone have an example of a document outlining "best practices" for how to prepare useful archived code (well annotated, etc) to go with data repositories?
— Daniel Bolnick (@DanielBolnick) November 21, 2021
This tweet by Daniel Bolnick received many useful replies with resources and guidelines for sharing code that’s useful and readable. Shortly after that, the editorial team at The American Naturalist published this guide guide for archiving data and code in ecology, evolution and behavior.
Coincidentally, I taught some relevant tools for this at a workshop last September and never wrote about it for this site. With a few packages and IDE tools, we can clean up our code efficiently, which goes a long way towards meeting the latest guidelines for clean, reproducible code.
Here’s a brief tour through my favorite tools:
packup
In interactive sessions and for less-structured workflows, I often add library() calls to my scripts after realizing I need a function without scrolling up to the top to put all the package load calls together. packup by Miles McBain provides an Rstudio addin to move all these calls up to the top of the script, remove any duplicates, and sort them alphabetically. Packup libary() calls works for both .R and .Rmd files and I see no downside to calling it before sharing a script, just make sure that the reordering isn’t causing namespace conflicts.
With packup we can easily go from this:
# random script
library(readr)
dat <- read_csv("mydata.csv")
library(dplyr)
dat
library(janitor)
# fix letter case
dat %>% clean_names()to this:
# random script
library(dplyr)
library(janitor)
library(readr)
dat <- read_csv("mydata.csv")
dat
# fix letter case
dat %>% clean_names()Bonus tip: In RStudio use Alt + arrow keys to move whole lines of code up or down (I did to nudge the commented line to the very top of the text).
annotater
The functions and RStudio addins in annotater are only a small step towards reproducibility (for the real deal, I suggest renv). After some time using them, I think they’ve proven their worth. Whenever I share code (including for the posts on this site, I use the Annotate package repository sources in active file addin to automatically annotate my library load calls with the source of the packages being loaded (e.g. CRAN, GitHub, BioConductor, etc.) and the version number.
annotater also has a cool function that can make a note of which functions are called from each of the packages being loaded in a file with ‘library’ calls. This can be useful to avoid dependency issues or making others install packages they don’t need (for running the code in that particular file).
Use the Annotate package repository sources in active file function to turn this:
# random script
library(dplyr)
library(janitor)
library(readr)into this:
# random script
library(dplyr) # CRAN v1.0.7
library(janitor) # CRAN v2.1.0
library(readr) # CRAN v2.0.2styler
styler can help us format our code consistently using a style guide that generally leads to cleaner code that is easier to read (e.g., correct indentation, spaces after commas and around infix operators but not after opening parenthesis for function calls, etc.). We can style a selection or an entire file.
This mangled code has some spacing and style issues:
ggplot( cast)+ geom_point(aes
(cost_gold,build_time, fill=
hit_points),pch=21,color="black", size=4)+
scale_fill_scico(
palette = "davos",direction = -1)+theme_clean(base_family = "Lato" )styler functions can help with that, returning the code below. Note that I normally don’t use the built-in tidyverse style guide (derived from the Google Style guide for R code) for ggplot code (too many line breaks after opening parens).
ggplot(cast) +
geom_point(aes
(cost_gold, build_time,
fill =
hit_points
), pch = 21, color = "black", size = 4) +
scale_fill_scico(
palette = "davos", direction = -1
) +
theme_clean(base_family = "Lato")Code sections
RStudio (and other IDEs) let us insert foldable sections, to split up scripts into discrete pieces that can be collapsed and navigated between using the little navigation panel that can be toggled on and off in the source pane. These can be built automatically for any comment with four trailing dashes, like so:
# My section ---- becomes
# My section ---------------------------------------- We can also insert these sections with Ctrl+Shift+R (Cmd+Shift+R on a Mac), and for longer scripts they bring more sanity.
littleboxes
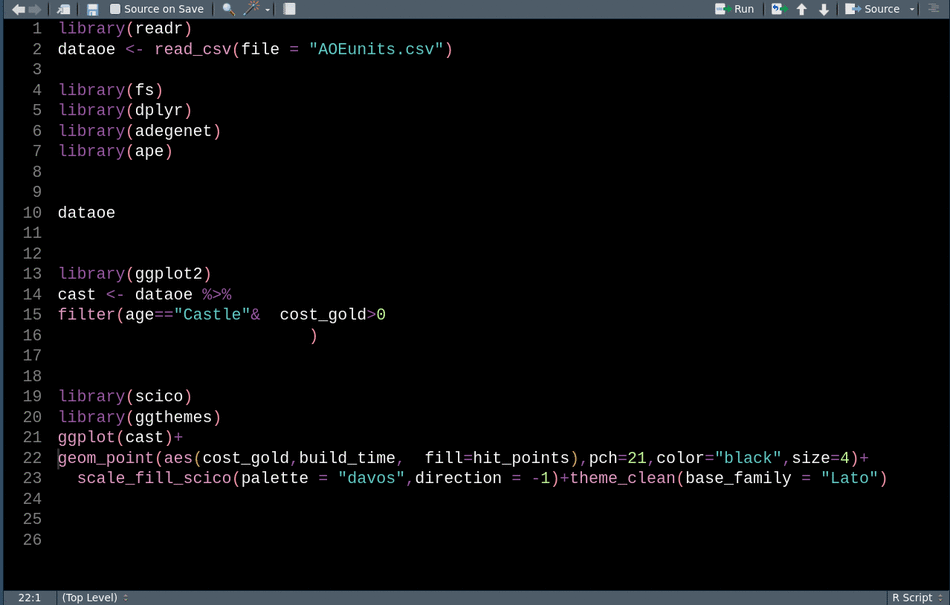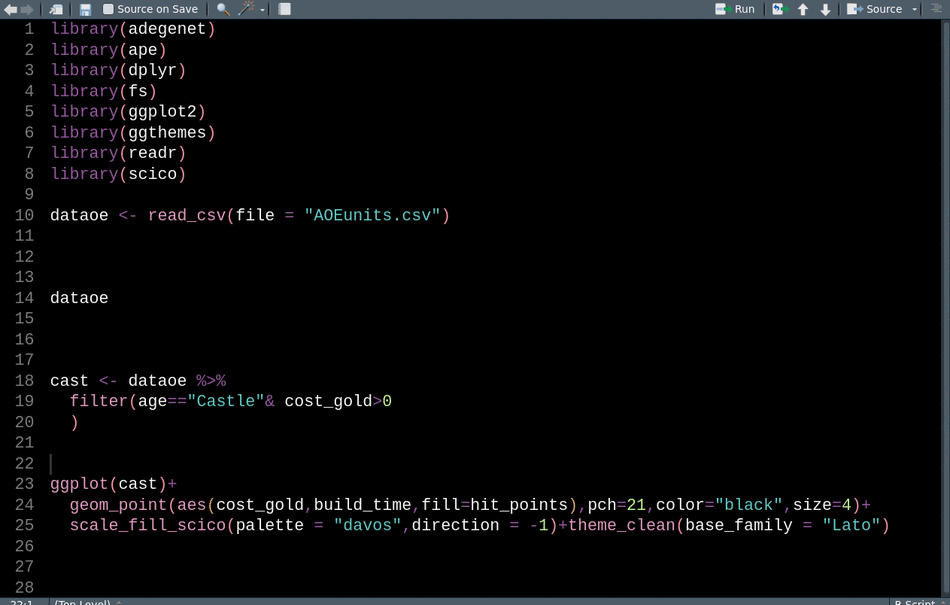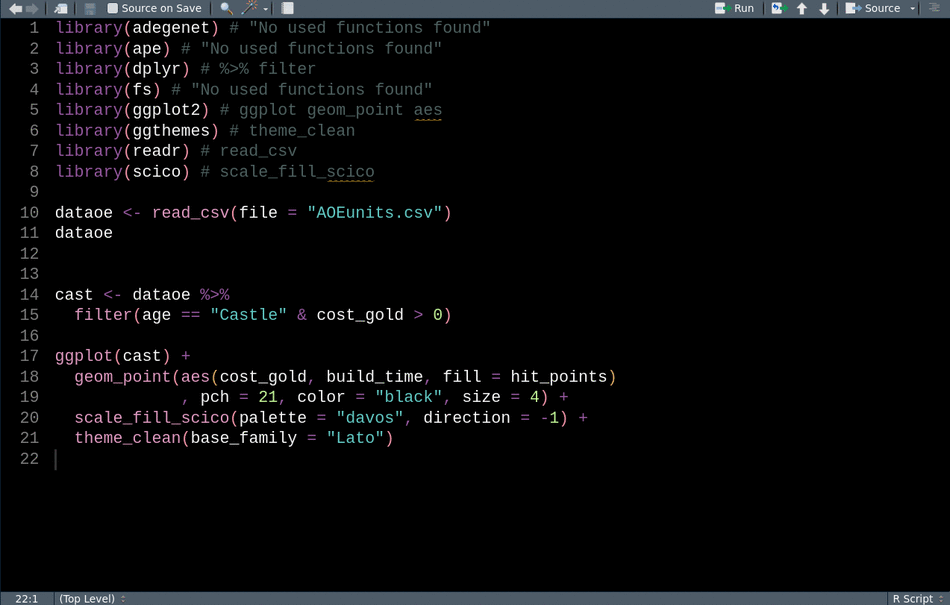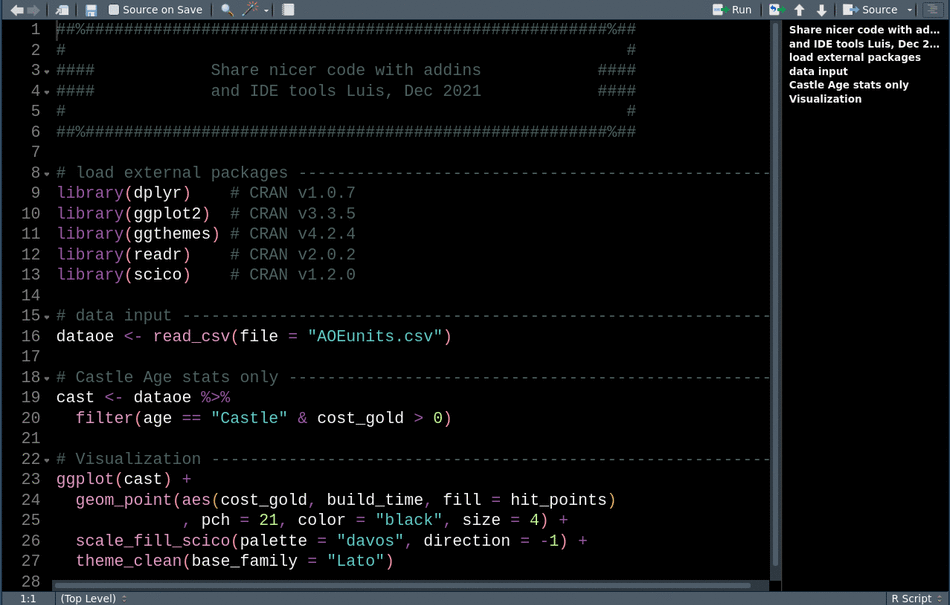
Lastly, littleboxes by the ThinkR squad gives us an addin for creating text titles with fancy ascii art boxes around them. I like to use these at the beginning of scripts, to include a date, the purpose of the file, and the author.
With the example from earlier, we can add titles, comments, dates or whatever, then select these lines and call Little Boxes to add a fancy frame.
This:
# Random script - Demonstrating IDE tools
#Dec 2021. by Luis
library(dplyr) # CRAN v1.0.7
library(janitor) # CRAN v2.1.0
library(readr) # CRAN v2.0.2Becomes this:
##%######################################################%##
# #
#### Random script - Demonstrating IDE ####
#### tools Dec 2021. by Luis ####
# #
##%######################################################%##
library(dplyr) # CRAN v1.0.7
library(janitor) # CRAN v2.1.0
library(readr) # CRAN v2.0.2This animation shows how we can use these tools sequentially to clean up our code for sharing.
Try these tools out!
