Tidying formatted spreadsheets with unheadr
unheadr
0.2.1 is now on CRAN, so here is an updated example for using the two main functions in the package.
The screenshot below comes from a spreadsheet with data from a regional event for the American Kennel Club’s Coursing Ability Test (CAT), which consists of timed 100-yard dashes for dogs. This is a subsample of an original xlsx file that I found online at some point and randomized.
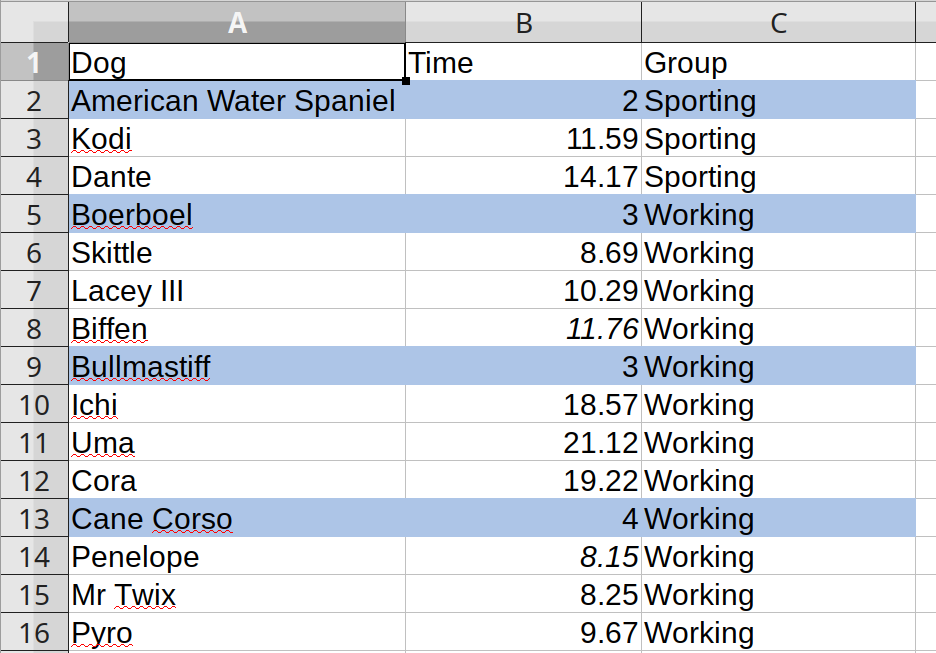
The breeds are embedded in the same column (variable) that holds the names of the individual dogs. Fortunately, the rows with the breeds have cell highlighting. Otherwise there would be no easy way to identify the breed names without prior knowledge (e.g. is Vizsla a name or a breed?). These ‘embedded subheaders’ approach is not quite tidy, and it would not be easy to, for example, calculate the average time by breed.
To work with these data in R, we can use the readxl package to import the xlsx file.
library(dplyr) # A Grammar of Data Manipulation
library(unheadr) # Handle Data with Messy Header Rows and Broken Values
# get the spreadsheet
CATspsheet <- tempfile(fileext = ".xlsx")
curl::curl_download("https://github.com/luisDVA/codeluis/blob/master/fastCAT2015.xlsx?raw=true", CATspsheet)
# import with readxl
readxl::read_excel(CATspsheet)Let’s have a look at the ouput from readxl::read_excel().
# A tibble: 32 x 3
Dog Time Group
<chr> <dbl> <chr>
1 American Water Spaniel 2 Sporting
2 Kodi 11.6 Sporting
3 Dante 14.2 Sporting
4 Boerboel 3 Working
5 Skittle 8.69 Working
6 Lacey III 10.3 Working
7 Biffen 11.8 Working
8 Bullmastiff 3 Working
9 Ichi 18.6 Working
10 Uma 21.1 Working
# … with 22 more rowsWith unheadr, we can turn the spreadsheet formatting (meaningful formatting) into character strings and then work with that.
# import and annotate
breeds_dat <- annotate_mf(CATspsheet, orig = Dog, new = dog_annotated)
breeds_datApplying the annotate_mf() function to annotate the Dogs variable, which adds a new variable that tells us about the type of formatting in each cell. The cell highlighting and HTML color code are appended onto the variable of interest.
# A tibble: 32 x 4
Dog dog_annotated Time Group
<chr> <chr> <dbl> <chr>
1 American Water Spaniel (highlighted-FFADC5E7) American Water Spaniel 2 Sporting
2 Kodi Kodi 11.6 Sporting
3 Dante Dante 14.2 Sporting
4 Boerboel (highlighted-FFADC5E7) Boerboel 3 Working
5 Skittle Skittle 8.69 Working
6 Lacey III Lacey III 10.3 Working
7 Biffen Biffen 11.8 Working
8 Bullmastiff (highlighted-FFADC5E7) Bullmastiff 3 Working
9 Ichi Ichi 18.6 Working
10 Uma Uma 21.1 Working
# … with 22 more rowsWe can then match the annotation strings with regex, and put the breed names into their own variable, leading to a tidy rectangular table (after some minor cleanup).
# match the rows with breed names and clean up
breeds_dat <- breeds_dat %>%
untangle2("^\\(", orig = dog_annotated, new = breed) %>%
mutate(breed = stringr::str_remove(breed, "^\\(.+\\)\\s")) %>%
select(-dog_annotated)Tidiness
# A tibble: 24 x 4
Dog Time Group breed
<chr> <dbl> <chr> <chr>
1 Kodi 11.6 Sporting American Water Spaniel
2 Dante 14.2 Sporting American Water Spaniel
3 Skittle 8.69 Working Boerboel
4 Lacey III 10.3 Working Boerboel
5 Biffen 11.8 Working Boerboel
6 Ichi 18.6 Working Bullmastiff
7 Uma 21.1 Working Bullmastiff
8 Cora 19.2 Working Bullmastiff
9 Penelope 8.15 Working Cane Corso
10 Mr Twix 8.25 Working Cane Corso
# … with 14 more rowsOn the event website, I later learned that italic text in the Time variable indicates that a dog was disqualified (for whatever reason). We can repeat the previous steps with the times variable, and join the resulting tables.
# repeat with Time variable
times_dat <- annotate_mf("pup/fastCAT2015.xlsx", orig = Time, new = time_annotated)
times_annotated <- times_dat %>% mutate(disqualified = if_else(stringr::str_detect(time_annotated, "italic"), TRUE, FALSE))
left_join(breeds_dat, times_annotated) %>% select(-time_annotated)# A tibble: 24 x 5
Dog Time Group breed disqualified
<chr> <dbl> <chr> <chr> <lgl>
1 Kodi 11.6 Sporting American Water Spaniel FALSE
2 Dante 14.2 Sporting American Water Spaniel FALSE
3 Skittle 8.69 Working Boerboel FALSE
4 Lacey III 10.3 Working Boerboel FALSE
5 Biffen 11.8 Working Boerboel TRUE
6 Ichi 18.6 Working Bullmastiff FALSE
7 Uma 21.1 Working Bullmastiff FALSE
8 Cora 19.2 Working Bullmastiff FALSE
9 Penelope 8.15 Working Cane Corso TRUE
10 Mr Twix 8.25 Working Cane Corso FALSE
# … with 14 more rowsPretty cool!
Writing this post made me realize that I should probably rewrite annotate_mf() so that we can specify if we need to skip rows when importing spreadsheets, and also think about a version of the function that can be applied across multiple variables simultaneously. If anyone wants to contribute please reach out.

