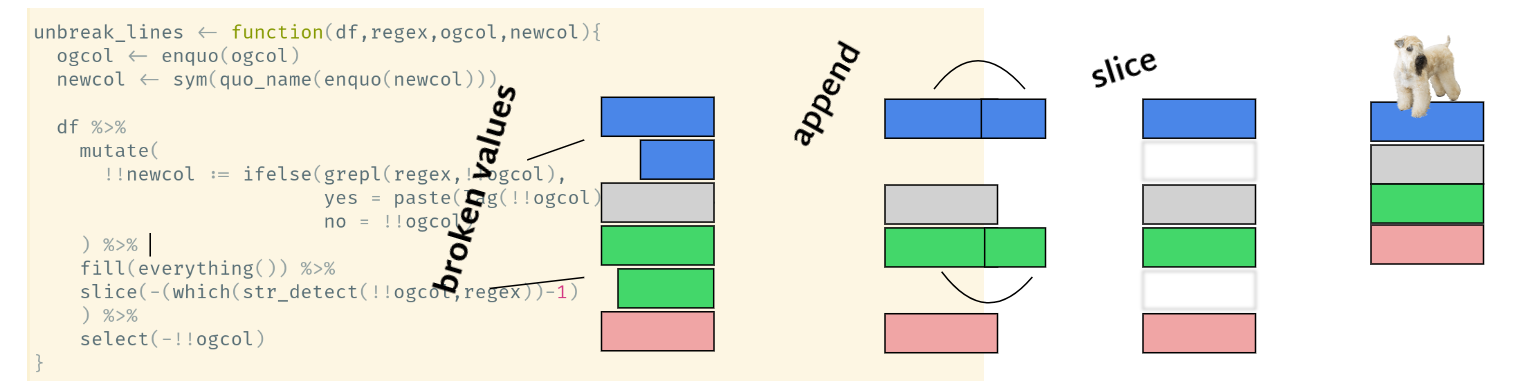
Tidyeval meets PDF table hell
Update - April 2018 - In the most recent release of rlang (0.2.0), we can use ensym() as a new variant of enexpr() for cleaner code. I’ve updated the code to reflect this change. Thanks to Hadley Wickham for the heads up.
Although it first became a feature of dplyr in June of 2017, tidy evaluation is once again in the spotlight after the 2018 RStudio conference. This is a good compilation of tidyeval resources, and I suggest watching this five-minute video of Hadley Wickham explaining the big ideas behind tidy evaluation while wearing a stylish sweater.
When tidyeval originally came out, I jumped at the chance to program with dplyr. I blogged about writing a function to deal with non-data rows embedded as hierarchical headers in the data rectangle. Unsurprisingly, I butchered the use of tidyeval and function writing in general, but I was rescued by Jenny Bryan in this post.
As a biologist, the ‘untangle’ function that came out of that exchange has saved me hours upon hours of work, because comparative data always has taxonomic header rows that I usually had to tidy up by hand in a spreadsheet program.
PDF table hell
In my ongoing work with other people’s data, I came across values that are broken up into two lines for whatever reason (often to optimize space on a page in a table in a typeset pdf).
I encounter broken-up values frequently in my biology research, here’s an example that isn’t made up.
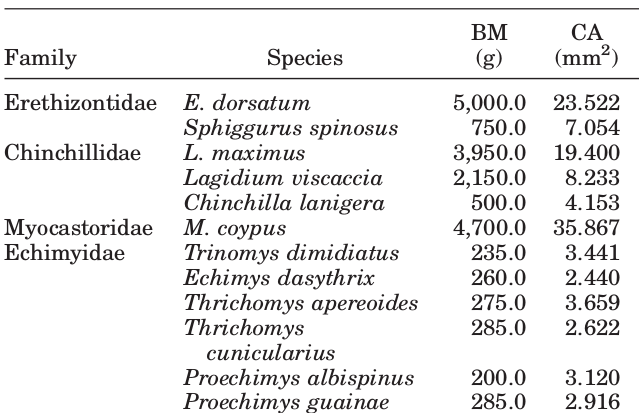
This is a very common practice, a lot of the pdf tables that I work with (using the awesome tabulizer package) have ‘merged’ cells that end up as broken values.
Here’s a toy example with some data from the summer Olympics.
| Games | Country | Soccer_gold_medal |
|---|---|---|
| Los Angeles 1984 | USA | France |
| Barcelona | Spain | Spain |
| 1992 | NA | NA |
| Atlanta 1996 | USA | Nigeria |
| Sydney 2000 | Australia | Cameroon |
| London | UK | Mexico |
| 2012 | NA | NA |
The values for two of the games (Barcelona 1992 & London 2012) are broken up into separate rows, adding a bunch of empty/NA values in the rows that shouldn’t really be there.
This is what the table should look like:
| Games_unbroken | Country | Soccer_gold_medal |
|---|---|---|
| Los Angeles 1984 | USA | France |
| Barcelona 1992 | Spain | Spain |
| Atlanta 1996 | USA | Nigeria |
| Sydney 2000 | Australia | Cameroon |
| London 2012 | UK | Mexico |
Using Jenny Bryan’s version of the untangle function as a template, I wrote the ‘unbreak_vals’ function below to unbreak values using tidyeval.
Assuming that:
the NA values in the table only correspond to the rows with broken-up values
the broken-up values can be matched with regex
this function will glue the two value fragments together and get rid of the extra row (via a hacky fill-then-slice operation).
Let’s try it out.
After loading the tidyverse set of packages and rlang, we’ll create the above table, define the “unbreak_vals” function, and use it – matching the rows that start out with numbers with the regex.
library(tidyverse)
library(rlang)
OGames <- tibble(Games = c("Los Angeles 1984","Barcelona","1992","Atlanta 1996","Sydney 2000","London","2012"),
Country = c("USA","Spain",NA,"USA","Australia","UK",NA),
Soccer_gold_medal = c("France","Spain",NA,"Nigeria","Cameroon","Mexico",NA))Let’s check it out
> OGames
# A tibble: 7 x 3
Games Country Soccer_gold_medal
<chr> <chr> <chr>
1 Los Angeles 1984 USA France
2 Barcelona Spain Spain
3 1992 NA NA
4 Atlanta 1996 USA Nigeria
5 Sydney 2000 Australia Cameroon
6 London UK Mexico
7 2012 NA NA Unbreak the lines, matching strings that start with a number
unbreak_vals <- function(df,regex,ogcol,newcol){
ogcol <- enquo(ogcol)
newcol <- ensym(newcol)
df %>%
mutate(
!!newcol := ifelse(grepl(regex,!!ogcol),
yes = paste(lag(!!ogcol),!!ogcol),
no = !!ogcol)
) %>%
fill(everything()) %>%
slice(-(which(str_detect(!!ogcol,regex))-1)
) %>%
select(-!!ogcol)
}
OGames %>% unbreak_vals("^[0-9]",Games,Games_unbroken) %>%
select(Games_unbroken,everything())It worked!
A tibble: 5 x 3
Games_unbroken Country Soccer_gold_medal
<chr> <chr> <chr>
1 Los Angeles 1984 USA France
2 Barcelona 1992 Spain Spain
3 Atlanta 1996 USA Nigeria
4 Sydney 2000 Australia Cameroon
5 London 2012 UK Mexico Another case of broken values that I’ve seen is when additional descriptions are interspersed below the original values in separate rows. This is a single-column example from a spreadsheet I had lying around.
dogsDesc <- tibble(dogs=c("Terrier","(Lakeland)","Terrier","(Soft-coated wheaten)","Bulldog","(English)","Bulldog","(French)"))> dogsDesc
# A tibble: 8 x 1
dogs
<chr>
1 Terrier
2 (Lakeland)
3 Terrier
4 (Soft-coated wheaten)
5 Bulldog
6 (English)
7 Bulldog
8 (French) Matching the opening bracket with the regex:
dogsDesc %>% unbreak_vals("^\\(",dogs,dogs_desc)# A tibble: 4 x 1
dogs_desc
<chr>
1 Terrier (Lakeland)
2 Terrier (Soft-coated wheaten)
3 Bulldog (English)
4 Bulldog (French) I have lots to learn about writing functions, but so far this ‘unbreak_vals’ function has already saved me lots of time and hassle and painful spreadsheet editing. If you have any questions or if you find this helpful please let me know.